CS 385 : Medical Imaging
Introduction
The goal of our project was to create an image classification system various types of Interstitial Lung Disease (ILD), including emphysema, ground-glass, micronodules, and fibrosis. Our system would take in CT scans of various patients and determine whether a given image shows healthy lungs or those with one of the mentioned diseases. We started by choosing our dataset from Talisman Test Suite, which contains over 18,000 CT scans from 97 different patients.
We decided early on that we would use our previous Assignment 3 code as a base for our classification system. The code already included SIFT feature extractors and a binary SVM approach to classification. Since there were multiple components to the code that needed to be updated/replaced, we narrowed down our ambitions for the project into 3 categories:
- Reformatting Images and Making the Code Image-Compatible
- Adding Multi-class Support Vector Machines
- Replace SIFT With Another Texture Extraction Technique
Our Approach & Algorithm
Reformatting Images & Codebase
The pixels from our CT scans had a much wider range of values compared to what Matlab can handle. Thankfully, we were able to convert these images fairly easily to RGB/Grayscale images fairly easily using a python script from a previous student. Next, we had to reformat the file structure for our images and change how our code would handle these new directories. This was necessary for getting our multi-class SVM to run down the road. This became particularly important because we started to notice a bias in our testing method. Our code was set to randomly select images, however most of our testing images came from the same patients as our training images. We chose a "leave one out" approach to our testing/training image selection process. This method selects one patient to test on, while training on the images from all other patients. This approach also allowed us to expand capabilities later and implement a full "leave one out" or k-fold Cross Validation.


Converting our CT scans (.tif) to Grayscale(.jpg)

K-folds Cross Validation
Multi-Class SVM
One of the most important aspects to our project was upgrading our binary SVM classifier to one that could handle multiple classes. Without a method to differentiate between our multiple classes, our systemís functionality would be minimal. Matlab provided great documentation on how to use our existing binary classifier for multiple classes using fitcsvm(). While we were able to run the classifier on some example Matlab code, we were not able to reorganize our directories for our system's testing.
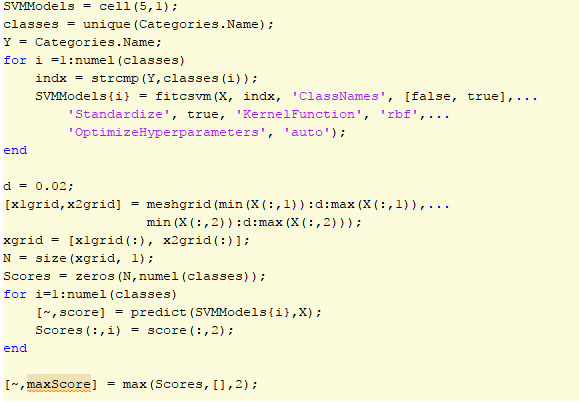
Matlab code used for Multi-class SVM
Filter Banks & Textons
Choosing a method for texture extraction turned out being the hardest part of the project because we were not able to run most of the online resources we found with our pre-existing code. Originally, we researched filter banks and texton forests because they proved to be the most accurate and efficient solution to our problem. While applying filter banks to our images gives a large amount of responses (approximately 39 responses for each image), special filters and texton decision forests would reduce the dimensionality of our feature vectors. We decided that the MRS4 filter would be the most efficient for our purposes, but any MR filters would also be acceptable to use.


Results
In our preliminary testing, we obtained surprisingly accurate results with binary classification. This happened because most of the images that we tested and trained on were obtained from the same patients. This caused a certain bias in our classifier that would make any future results invalid. Unfortunately, we weren't able to get many results otherwise. Our team fell far short from what we expected to achieve over the course of our project. While we were most excited about the new use of filter banks and texton decision forests, we spent most of our time getting the pre-existing code to handle our multiple classes. Getting our code running and working correctly ended up being our primary priority in the entire second sprint, pulling our attention away from texture extraction methods. As far as what we were able to accomplish, we were able to pre-process and classify our images with SIFT and multi-class SVM. We were also able to generate some simple RFS filters, but we could not reduce the dimensionality down to our target MRS4, MR4 or MR8 filters. These filters didnít lead anywhere because we did not implement texton forests at all.
Conclusions
In retrospect, thereís many things we would have done differently looking back at our decisions. The codebase that we chose to build on top of is our first regret because, although organized, the Assignment 3 code was unnecessarily large for our purpose. Whenever changes were made to one step in the codeís pipeline, it had multiple implications and effects on the rest of the codebase. 90% of our time was spent debugging what errors our changes had introduced. Creating our own codebase and selecting the functions and files that we needed would have proved much easier and less time-consuming. We also heavily underestimated the complexity of some of the methods we wanted to use in our project, specifically filter banks and texton forests. Finally, our inexperience in Matlab caused many problems. Some online resources, and even our original Assignment 3 code, used files other than Matlab scripts (.m files) to run. The files also held dependencies on the operating system being used, so moving back and forth from our windows machines to the lab computers proved to be tough.