CS 385: Animal Classification
Introduction:
What we were aiming to achieve:
- Crop and preprocess images to reduce noise and obstruction.
- Finding if and where an animal is in a picture using template matching and a sliding window approach.
- Using bag of words, classify the animal in the picture.
Our group was able to obtain about 700 images of animals taken by the Sonoma State Center for Environmental Inquiry. We started by going through the images and deciding on what categories of animals we could possibly identify with our program and cropping images for testing. We used this data to classify and identify animals, with template matching and bag of words methods. We wanted to be able to tell if there was an animal in an image, what kinds of animal it is and where the animal is in the image. We decided to use bag of words for our classifer since we already had some experience with it. For object detection, you recommended we use template matching for detecting the animals and we were able to find a video to get us going (see references). Depending on the templates we used, we had varying degrees of success. Templates that were heavily saturated with light pixels (overexposed) lead to false positive results, while templates with more contrast matched better. Bag of words was used to classify the animals and much more accurate at classifying than template matching.
Algorithm/Approach:
Template Matching Approach: This process is pretty simple, we first crop the images so that we have a full size image, and a small cropped image of just the animal. We attempmted to do some preprocessing on the images, like unsharp masking and applying a median filter, but preprocessing all the images was time consuming and yielded too little improvement over processing time. We then need to convert the images to grayscale images to use with the normxcorr2 function. We use the normxcorr2 to compute the Normalized Cross Correlation Matrix, this function drags the smaller image over the larger one (sliding box) and gives you a result based on how well the pixels match (0-1). We then find the max absolute value in the Cross Correlation matrix and that should be where the animal is (given a template with matching contrast and position). Finaly we draw a bounding box around where we think the animal should be and write the classification. The code for template mathching is shown below:
Template Matching Code:
% Read cropped image
CroppedImage = imread(CroppedImageFile);
%convert cropped image to grayscale
CroppedImage = rgb2gray(CroppedImage);
% CI = cropped image
[RowCI, ColCI] = size(CroppedImage);
%sprintf('The size of the cropped image is %d%d.',RowCI, ColCI)
% Read target image
TargetImage = imread(TargetImageFile);
%convert target image to grayscale
TargetImage = rgb2gray(TargetImage);
% TI = target image
[RowTI, ColTI] = size(TargetImage);
%sprintf('The size of the target image is %d%d.',RowTI, ColTI)
% Calculate Normalized Cross Correlation Matrix
cc = normxcorr2(CroppedImage, TargetImage);
% Display the Normalized Cross Correlation Matrix
%imshow(cc), title('Normalized Cross Correlation Matrix')
% CC = Cross Correlation
[RowCC, ColCC] = size(cc);
%sprintf('The size of the Normalized Cross Correlation Matrix is %d%d.',RowCC, ColCC)
% Get the peak correlation value (max_cc) and its position (imax)
% Note imax is the position in a one column array
[max_cc, imax] = max(abs(cc(:)));
%sprintf('The highest correlation value is %f.', max_cc)
[ypeak, xpeak] = ind2sub(size(cc), imax(1));
%sprintf('The row col coords of the best match in the cross corr matrix is %d %d.',ypeak, xpeak)
BestRow = ypeak - (RowCI - 1);
BestCol = xpeak - (ColCI - 1);
%sprintf('the row col coords of the best match in orig matrix is %d %d.', BestCol, BestRow)
figure, imshow(CroppedImage);
figure, imshow(TargetImage);
% Use imrect to indicate the best match area
h = imrect(gca, [BestCol BestRow (ColCI - 1) (RowCI - 1)]);
H = text( BestCol, BestRow - 24, Animal);
set(H,'color','white','fontsize',22)
Bag of Words Approach: For a bag of words approach of the problem of classifying animals, we had to make modifications to the assignment 4 code to deal with multi-classification using SVM. The inherent issue with using SVM for multi-classification is that SVM is designed to only deal with a binary classification (the image is either one class, or it's not).
Bag of Words Code Snippet:
function highScore = animal_classification(config_file, im, preProccess)
eval(config_file)
codebook_name = [CODEBOOK_DIR , '\', VQ.Codebook_Type ,'_', num2str(VQ.Codebook_Size) , '.mat'];
load(codebook_name);
[imy,imx,imz] = size(im);
%%% Resize image, proved Preprocessing.Image_Size isn't zero
%%% in which case, do nothing.
if (Preprocessing.Image_Size>0)
%%% Figure out scale factor for resizing along appropriate axis
if strcmp(Preprocessing.Axis_For_Resizing,'x')
scale_factor = Preprocessing.Image_Size / imx;
elseif strcmp(Preprocessing.Axis_For_Resizing,'y')
scale_factor = Preprocessing.Image_Size / imy;
else
error('Unknown axis');
end
%%% Rescale image using bilinear scaling
im = imresize(im,scale_factor,Preprocessing.Rescale_Mode);
else
scale_factor = 1;
end
%do sift on probe image
[row col depth] = size(im);
%convert to grayscale
if depth > 1
im = rgb2gray(im);
end
%apply median filter to image and then sharpen
im = medfilt2(im, [3 3]);
im = imsharpen(im);
% Find Interest points using vl_sift. Store features in f and descriptors in d.
[f,d] = vl_sift(single(im));
% Total number of features from image
nFeats = size(f,2);
% From f and d extract x, y, scale, angle and descriptor.
x = f(1,:);
y = f(2,:);
scale = f(3,:);
angle = f(4,:);
descriptor = d;
%create histogram of features for current image
nImgPoints = length(scale);
distance = Inf * ones(nImgPoints, VQ.Codebook_Size);
for s = 1:nImgPoints
for q = 1:VQ.Codebook_Size
distance(s, q) = norm(centers(:,q) - double(descriptor(:,s)));
end
end
[temp, descriptor_vq] = min(distance, [], 2);
hist = zeros(1, VQ.Codebook_Size);
for s = 1:nImgPoints
hist(descriptor_vq(s)) = hist(descriptor_vq(s))+1;
end
% fprintf('Number of features detected for probe image: %d\n',length(x));
fprintf('.');
load([RUN_DIR,'\completeAnimalSVM.mat']);
%determine number of classes to be trained for 1 vs all method
cat_length = size(Categories.Name, 1)-10;
bgIndexStart = cat_length+1;
bgIndexEnd =cat_length+10;
%contains an index and its score
highScore = [0 Inf];
for r = 1:cat_length
[prediction score] = predict(AnimalSVM{r}{1,1}, hist);
% if classified check score against back ground classification, if
% score > than positive background identification score, compare
% score to known high scores already detected, if larger, update
% highscore
if score(:,1) > AnimalThresh(r)
%asssum score is higher
higherScore = 1;
%iterate through backgorund classifications
for s = bgIndexStart:bgIndexEnd
%compare current windows histogram to current background
%class
[temp bg_score] = predict(AnimalSVM{s}{1,1}, hist);
%if the bg_score is greater than the thresh for current
%background compare that score to the current class score
if bg_score(1) > AnimalThresh(s)
if score(:,1) < bg_score(:,1)
higherScore = 0;
end
end
end
%current classifcation from class index r has higher confidence
%than background score
if higherScore == 1
%if current highscore isn't present set high score to current
%score
if isinf(highScore(2))
highScore = [r score(:, 1)];
% if current score is higher than previous score, set current
% score to high score
elseif score(:,1) > highScore(2)
highScore = [r score(:,1)];
end
end
end
end
To overcome this issue To overcome this issue we had to use a one versus all (a.k.a. One versus rest) approach. The idea behind one versus all is to separate one class from all other classes. In other words, either an image has that one species or it’s not that species. So for each class of animal, we had to create an SVM model and store it in an array that is 1xn (where n is the number of classifiers being tested). This allows for quick processing of probing images that are being checked for any animals.
For testing probe images, we used a sliding window method to get higher detail for feature detection. For each window image, we had to perform a sift operation on the image and extract the features from the image. From there a histogram of the features needed be created using the codebook created when training the classifiers. This was done by comparing each feature from the probing image to each individual cluster in the codebook and determining which cluster the feature is closest to. The result of this comparison is a kx1 matrix (where k = the number of clusters in the codebook).
After creating the histogram of features, we were required to use that histogram and each SVM created during the training phase to predict whether or not that window image has a particular animal in that window. We also had to track the highest score for any image that was greater than the ROC threshold as we operated under the assumption that the highest score indicated the confidence of that animal.

Results
Intermediate Results:


Some of the bad results we got from bag of words:


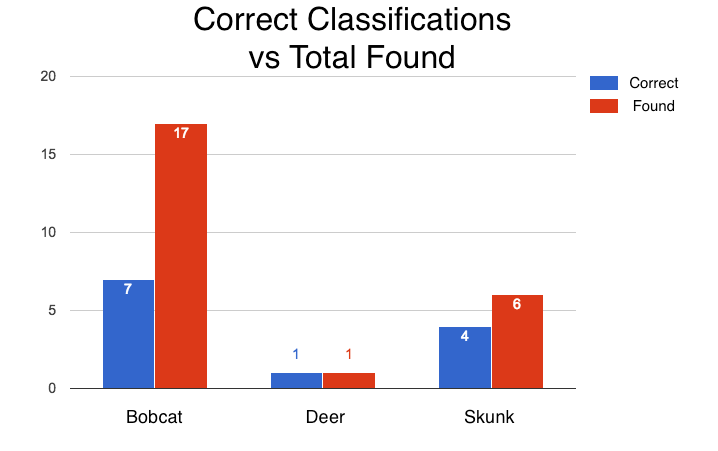
Bag of Words Results: Considering the poor quality of the images collected, we had poor success rates using bag of words for classification. While we tried several different methods to reduce false positives (e.g. breaking background training images into smaller quadrants) our results were mostly poor. This lead to us having to reduce the number of animal classifications to just two; deer and skunk. We chose these two classes because they had the “clearest” images. This adjustment helped abate some misclassification, but there were still issues with misclassification of the background.
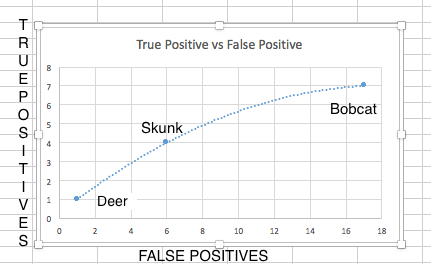
Template Matching Results: Template Matching excelled at finding bounding boxes in using a cropped version of the image. Using the high correlation values from the cropped images we can manually classify the images. Template Matching had problems though, it would incorrectly classify parts of the background as bobcats. Classifying and creating bounding boxes for blurry images incorrectly as well.
Discussion and Conclusions:
The choice to go for nighttime pictures turned out to be more of a challenge than we anticipated. A large amount of the nighttime images were overexposed and obscured by bright plants in the foreground. The bright plants were focused on by our descriptors and overexposed the rest of the image (causing some of the features on the animals to be lost). Template matching is very naive approach for this problem. Templates with poor contrast lead to false positive results, (bonding box not around the animal). Also we needed to make templates for each different position of the animals to be able to detect them.
References:
- Template Matching code
- Bag of Words starter code from Homework 4